2022 Blogs, Blog, Featured
Genomics Cloud on AWS with RLCatalyst Research Gateway
The pandemic worldwide has highlighted the need for advancing human health faster and new drugs discovery advancement for precision medicines leveraging Genomics. We are building a Genomics Cloud on AWS leveraging convergence of Big Compute, Large Data Sets, AI/ML Analytics engines, and high-performance workflows to make drug discovery more efficient, combining cloud & open source with our products.
Relevance Lab (RL) has been collaborating with AWS Partnership teams over the last one year to create Genomics Cloud. This is one of the dominant use cases for scientific research in the cloud, driven by healthcare and life sciences groups exploring ways to make Genomics analysis better, faster, and cheaper so that researchers can focus on science and not complex infrastructure.
RL offers a product RLCatalyst Research Gateway that facilitates Scientific Research with easier access to big compute infrastructure, large data sets, powerful analytics tools, a secure research environment, and the ability to drive self-service research with tight cost and budget controls.
The top use cases for AWS Genomics in the Cloud are implemented by this product and provide an out-of-the-box solution, significantly saving cost and effort for customers.
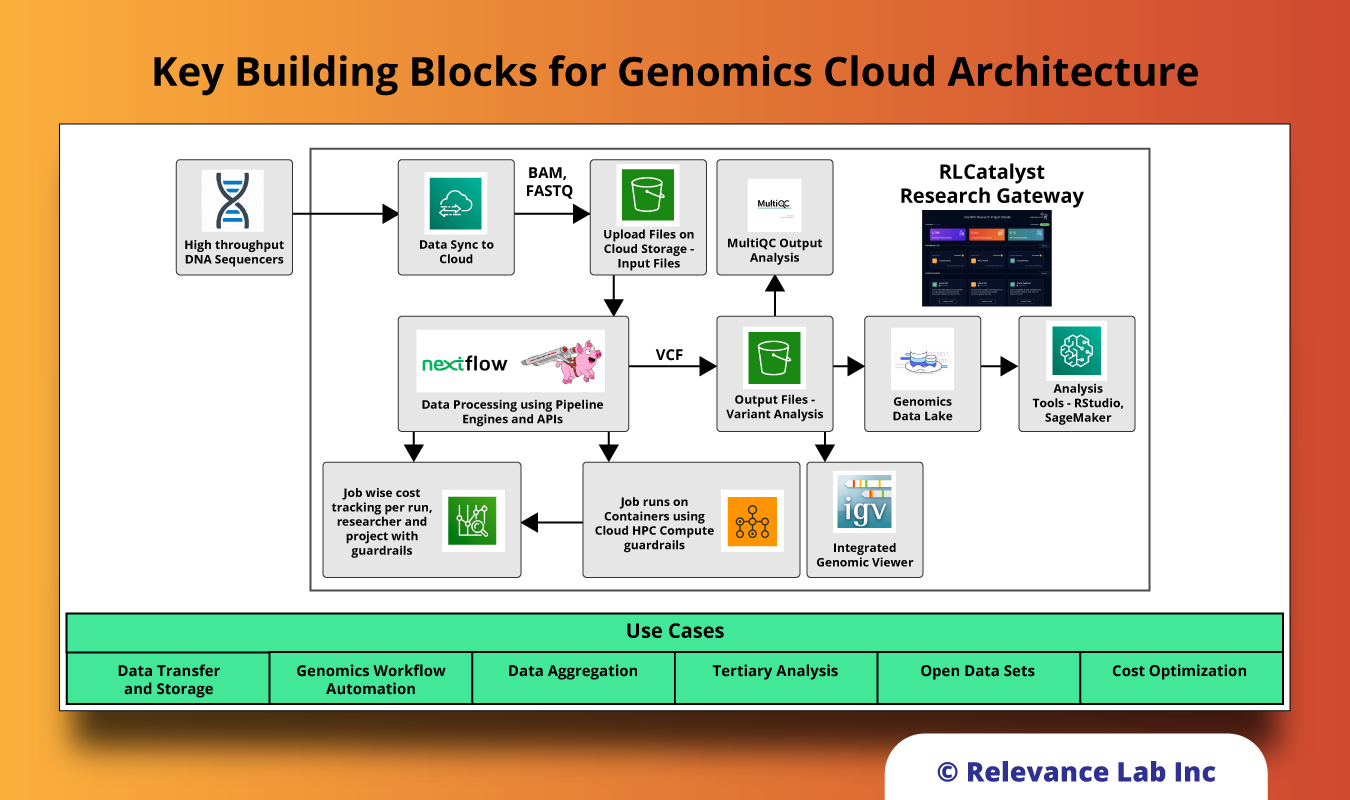
Key Building Blocks for Genomics Cloud Architecture
The solution for supporting easy use of Genomics Cloud supports the following key components to meet the need of researchers, scientists, developers, and analysts to efficiently run their experiments without the need for deep expertise in the backend computing capabilities.
Genomics Pipeline Processing Engine
The researchers’ community uses popular open-source tools like NextFlow and Cromwell for large data sets by leveraging HPC systems, and the orchestration layer is managed by tools like Nextflow and Cromwell.
Nextflow is a bioinformatics workflow manager that enables the development of portable and reproducible workflows. It supports deploying workflows on a variety of execution platforms, including local, HPC schedulers, AWS Batch, Google Cloud Life Sciences, and Kubernetes.
Cromwell is a workflow execution engine that simplifies the orchestration of computing tasks needed for Genomics analysis. Cromwell enables Genomics researchers, scientists, developers, and analysts to efficiently run their experiments without the need for deep expertise in the backend computing capabilities.
Many organizations also use commercial tools like Illumina DRAGEN and NVidia Parabricks for similar solutions that are more optimized in reducing processing timelines but also come with a price.
Open Source Repositories for Common Genomics Workflows
The solution needs to allow researchers to leverage work done by different communities and tools to reuse existing available workflows and containers easily. Researchers can leverage any of the existing pipelines & containers or can also create their own implementations by leveraging existing standards.
GATK4 is a Genome Analysis Toolkit for Variant Discovery in High-Throughput Sequencing Data. Developed in the Data Sciences Platform at the Broad Institute, the toolkit offers a wide variety of tools with a primary focus on variant discovery and genotyping. Its powerful processing engine and high-performance computing features make it capable of taking on projects of any size.
BioContainers – A community-driven project to create and manage bioinformatics software containers.
Dockstore – Dockstore is a free and open source platform for sharing reusable and scalable analytical tools and workflows. It’s developed by the Cancer Genome COLLABORATORY and used by the GA4GH.
nf-core Pipelines – A community effort to collect a curated set of analysis pipelines built using Nextflow.
Workflow Description Language (WDL) is a way to specify data processing workflows with a human-readable and -writeable syntax.
AWS Batch for High Performance Computing
AWS has many services that can be used for Genomics. In this solution, the core architecture is with AWS Batch, a managed service that is built on top of other AWS services, such as Amazon EC2 and Amazon Elastic Container Service (ECS). Also, proper security is provided with Roles via AWS Identity and Access Management (IAM), a service that helps you control who is authenticated (signed in) and authorized (has permissions) to use AWS resources.
Large Data Sets Storage and Access to Open Data Sets
AWS cloud is leveraged to deal with the needs of large data sets for storage, processing, and analytics using the following key products.
Amazon S3 for high-throughput data ingestion, cost-effective storage options, secure access, and efficient searching
AWS DataSync for secure, online service that automates and accelerates moving data between on premises and AWS storage services
AWS Open Datasets Program houses openly available, with 40+ open Life Sciences data repositories
Outputs Analysis and Monitoring Tools
One of the key building blocks for Genomic Data Analysis needs access to common tools like the following integrated into the solution.
MultiQC reports MultiQC searches a given directory for analysis logs and compiles an HTML report. It’s a general-use tool, perfect for summarising the output from numerous bioinformatics tools.
IGV (Integrative Genomics Viewer) is a high-performance, easy-to-use, interactive tool for the visual exploration of genomic data.
RStudio for Genomics since R is one of the most widely-used and powerful programming languages in bioinformatics. R especially shines where a variety of statistical tools are required (e.g., RNA-Seq, population Genomics, etc.) and in the generation of publication-quality graphs and figures.
Genomics Data Lake
AWS Data Lake for creating Genomics data lake for tertiary processing. Once the Secondary analysis generates outputs typically in Variant Calling Format (VCF) for further analysis, there is a need to move such data into a Genomics Data Lake for tertiary processing. Leveraging standard AWS tools and solution framework, a Genomics Data Lake is implemented and integrated with the end-to-end sequencing processing pipeline.
Variant Calling Format
specification is used in bioinformatics for storing gene sequence variations, typically in a compressed text file. According to the VCF specification, a VCF file has meta-information lines, a header line, and data lines. Compressed VCF files are indexed for fast data retrieval (random access) of variants from a range of positions.
VCF files, though popular in bioinformatics, are a mixed file type that includes a metadata header and a more structured table-like body. Converting VCF files into the Parquet format works excellently in distributed contexts like a Data Lake.
Cost Analysis of Workflows
One of the biggest concerns for users of Genomic Cloud is control on budget and cost that is provided by RLCatalyst Research Gateway by tracking spends across Projects, Researchers, Workflow runs at a granular level and allowing for optimizing spends by using techniques like Spot instances and on-demand compute. There are guardrails built-in for appropriate controls and corrective actions. Users can run sequencing workflows using their own AWS Accounts, allowing for transparent control and visibility.
Summary
To make large-scale genomic processing in the cloud easier for institutions, principal investigators, and researchers, we provide the fundamental building blocks for Genomics Cloud. The integrated product covers large data sets access, support for popular pipeline engines, access to open source pipelines & containers, AWS HPC environments, analytics tools, and cost tracking that takes away the pains of managing infrastructure, data, security, and costs to enable researchers to focus on science.
To know more about how you can start your Genomic Cloud in the AWS cloud in 30 minutes using our solution at https://research.rlcatalyst.com, feel free to contact marketing@relevancelab.com.
References
High-performance genetic datastore on AWS S3 using Parquet and Arrow
Parallelizing Genome Variant Analysis
Pipelining GATK with WDL and Cromwell
Accelerating Genomics and High Performance Computing on AWS with Relevance Lab Research Gateway Solution


