2022 Blogs, Blog, Featured
In recent times, Next Generation Sequencing (NGS) has transformed from being solely a
research tool to be routinely applied in many fields, including diagnostics, outbreak disease investigations, antimicrobial resistance, forensics, and food authenticity. The use of cloud and modern open source tools is helping advancement at a rapid pace, with continuous improvement in quality and cost reduction, and is having a major influence on food microbiology. Public health labs and food regulatory agencies globally are embracing Whole Genome Sequencing (WGS) as a revolutionary new method. In this blog, we try to introduce this interesting use case and cover a common use case of Bacterial Genome Analysis in the cloud using our Research Gateway. We will show how to run powerful tools like Bactopia, a Flexible Pipeline for Complete Analysis of Bacterial Genomes in a few minutes.
What is Bactopia?
Sequencing of bacterial genomes is gathering momentum for greater adoption. Bactopia, developed by Robert A. Petit III, was created with a new series of pipelines (acknowledgements) built using Nextflow workflow software to provide efficient comparative genomic analyses for bacterial species or genera. This pipeline has more advanced features than many others in a similar space.
The image below shows the High Level Components of Bactopia Pipeline.
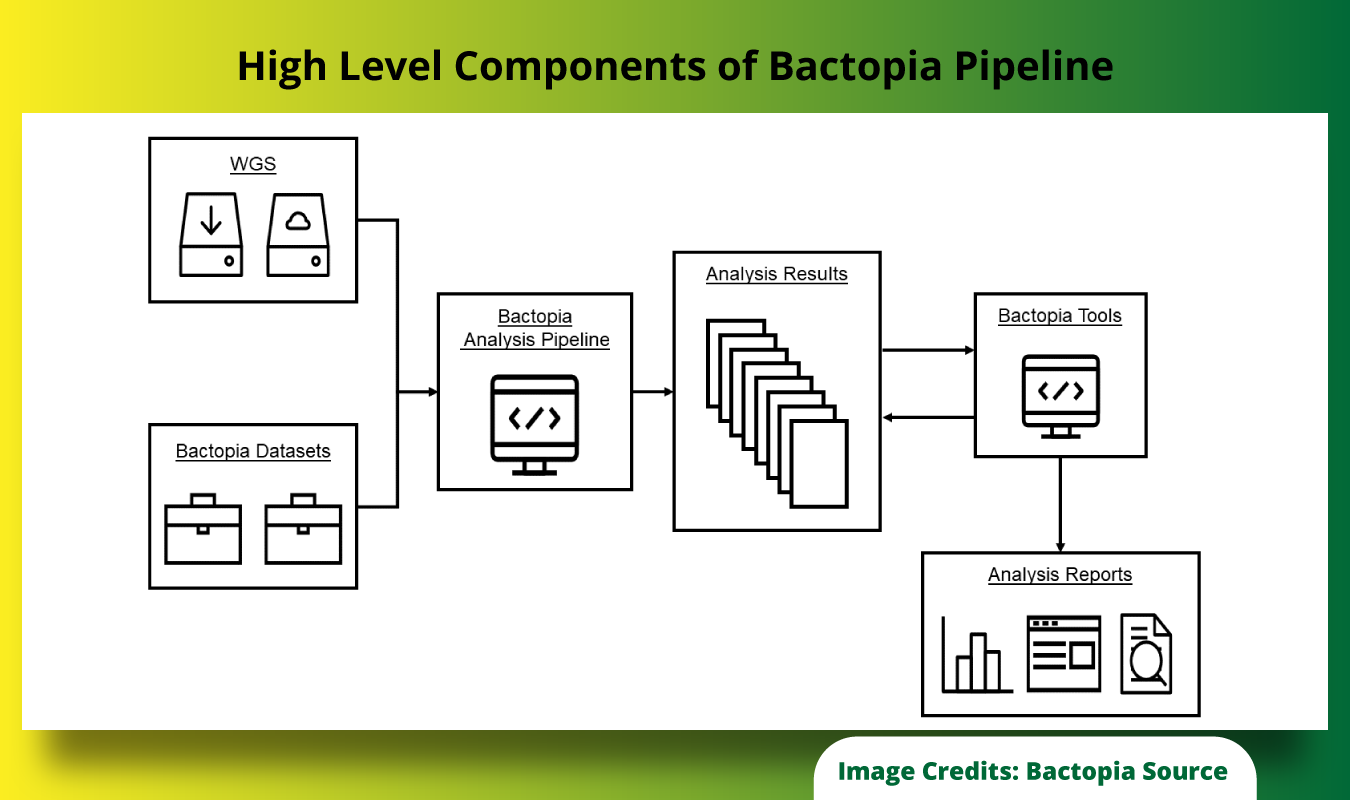
What Makes Bactopia More Powerful Compared to Other Similar Solutions?
The following data shared by the authors of this pipeline highlights the key strengths.
- List of bioinformatic tools used by the Bactopia Analysis Pipeline
- A comparison of bacterial genome analysis workflows
Usually, for researchers to get started with setting up a secure environment, accessing the data, big compute, and analytics tools can be a significant effort. With Research Gateway built on AWS, we make it extremely simple to get started.
An Introduction to Running Bactopia on AWS Cloud
Bactopia is a software pipeline for the complete analysis of bacterial genomes. Bactopia is based on the Nextflow bioinformatic workflow software. Research Gateway supports Nextflow based pipelines to be run with great ease, and we will show you how the same can be achieved with Bactopia.
Steps for Running Bactopia Pipeline on AWS Cloud
Step-1: Using the publicly available Bactopia repository on Github, a new AWS AMI is created by installing Bactopia software on Nextflow advanced product available as part of Research Gateway standard catalog. This step is needed since Bactopia contains a large number of specialized tools integrated and embeds Nextflow internally for its execution. Once the new AMI of Bactopia is ready, it is added to AWS Service Catalog and imported into Research Gateway to be used by Researchers. The product is available in the standard products category to be launched with 1-Click, as shown below.

Step-2: Once the Bactopia product is ordered using a simple screen as shown above in about 10 minutes, the setup with all the tools, Nextflow & Nextflow Tower are all provisioned and ready to be used. The user can log in to the Bactopia server using the SSH key-pair available from within the Portal UI using the “SSH/RDP Connect” action, as shown below.

Step-3: Copy data to the Bactopia server based on samples to be used for processing and start the execution of workflow as per available documentation. In our case, we tried with a smaller set of sample data sets, and it took us 15 min to run the pipeline and view outputs in the console window.
Step-4: When the pipeline is being executed using Nextflow Tower, details of the jobs and all key metrics can be viewed by the user from within the Research Gateway by selecting the “Monitor Pipeline” action. The entire complexity of different tools integrated on the platform is invisible to the user making it a seamless experience.
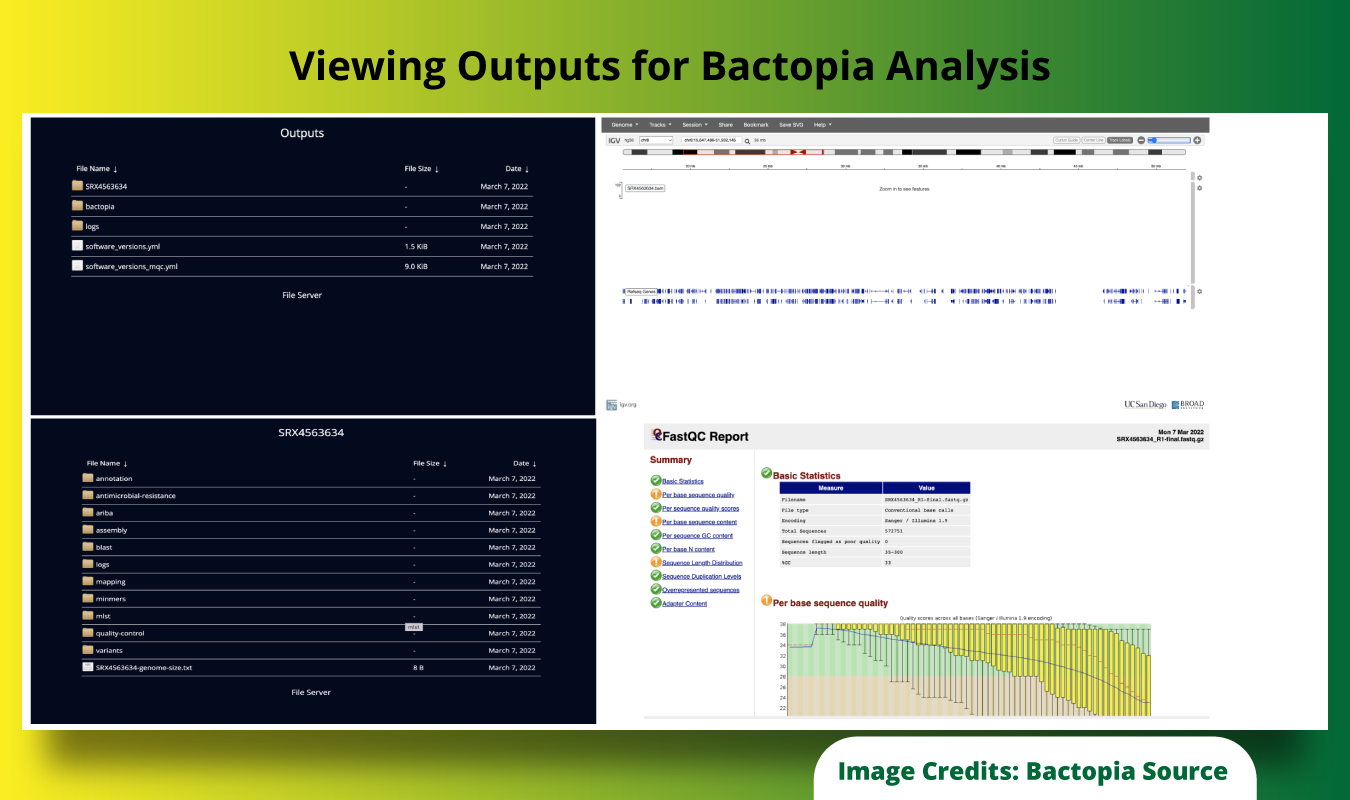
Step-5: The outputs generated by the Bactopia pipeline can be viewed from within the Portal using the “View Outputs” action that allows users to view the outputs in a simple browser, and actions can be taken to view the same with specialized tools like Integrative Genomics Viewer (IGV) or MultiQC reports, etc.
All the products that are used in Research Gateway have automatic tagging and tracking for cost purposes, and it can be easily verified by project, researchers, and product type on the total consumption providing a powerful cost management and budget tracking tool.
Summary
As the usage of Genomics adoption grows and new use cases emerge for leveraging the power of this technology, focus on food safety is a growing need with an ability to Bacterial Genome analysis using advanced pipelines popularly available in the open source community. To help researchers use such power tools with speed on the cloud without getting into the complexity of infrastructure, networks, security, and focus on science, we have demonstrated in this blog the ability to use Research Gateway to run your first pipeline in less than 60 minutes.
To know more about how you can start your Bacterial Genome analysis pipelines on the AWS Cloud in less than 60 minutes using our solution at https://research.rlcatalyst.com, feel free to contact marketing@relevancelab.com
References
An introduction to running Bactopia on Amazon Web Services (May 2021)
Using AWS Batch to process 67,000 genomes with Bactopia (December 2020)
Accelerating Genomics and High Performance Computing on AWS with Relevance Lab Research Gateway Solution
