2023 Blog, AWS Service, Research Gateway, Blog, Featured
Major advances are happening with the leverage of Cloud Technologies and large Open Data sets in the areas of Healthcare informatics that include sub-disciplines like Bioinformatics and Clinical Informatics. This is being rapidly adopted by Life Sciences and Healthcare institutions in commercial and public sector space. This domain has deep investments in scientific research and data analytics focussing on information, computation needs, and data acquisition techniques to optimize the acquisition, storage, retrieval, obfuscation, and secure use of information in health and biomedicine for evidence-based medicine and disease management.
In recent years, genomics and genetic data have emerged as an innovative areas of research that could potentially transform healthcare. The emerging trends are for personalized medicine, or precision medicine leveraging genomics. Early diagnosis of a disease can significantly increase the chances of successful treatment, and genomics can detect a disease long before symptoms present themselves. Many diseases, including cancers, are caused by alterations in our genes. Genomics can identify these alterations and search for them using an ever-growing number of genetic tests.
With AWS, genomics customers can dedicate more time and resources to science, speeding time to insights, achieving breakthrough research faster, and bringing life-saving products to market. AWS enables customers to innovate by making genomics data more accessible and useful. AWS delivers the breadth and depth of services to reduce the time between sequencing and interpretation, with secure and frictionless collaboration capabilities across multi-modal datasets. Also, you can choose the right tool for the job to get the best cost and performance at a global scale— accelerating the modern study of genomics.
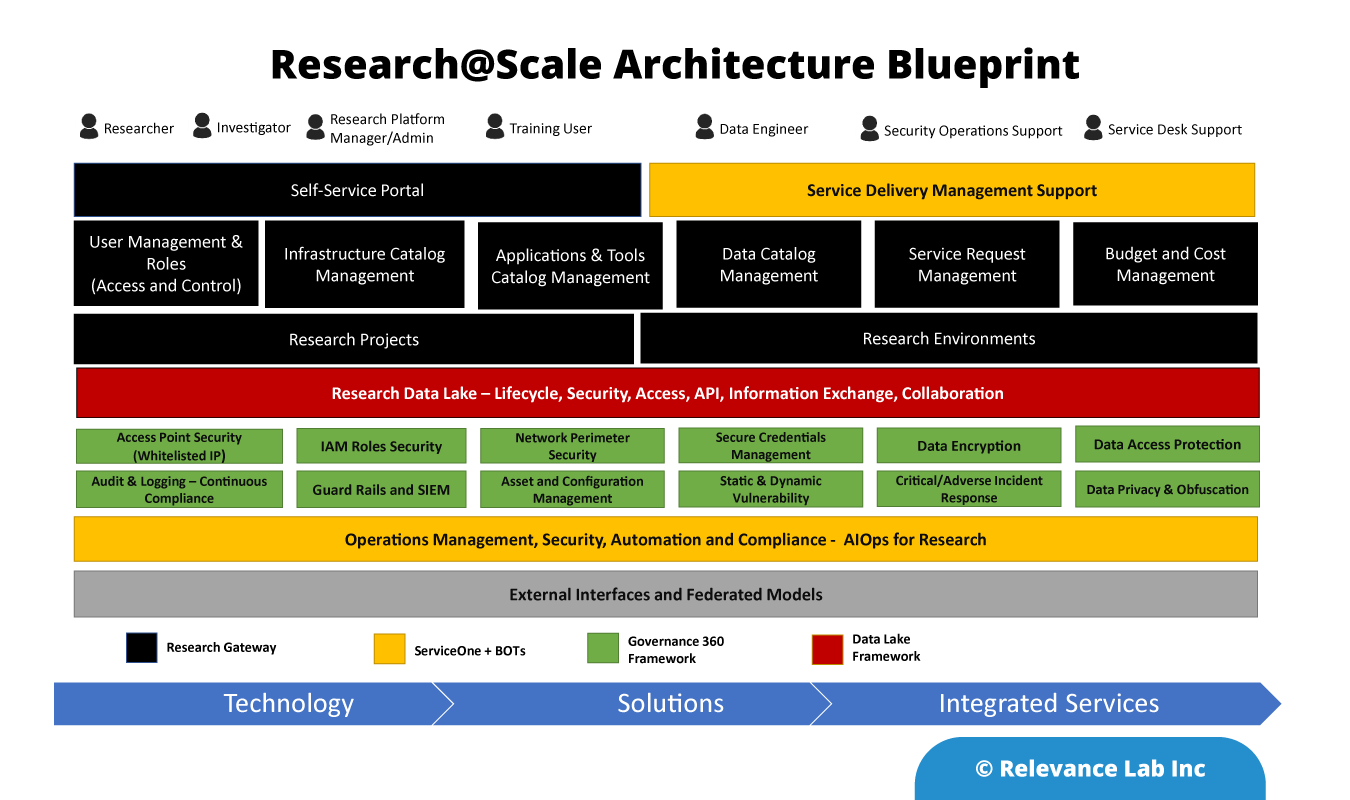
Relevance Lab Research@Scale Architecture Blueprint
Working closely with AWS Healthcare and Clinical Informatics teams, Relevance Lab is bringing a scalable, secure, and compliant solution for enterprises to pursue Research@Scale on Cloud for intramural and extramural needs. The diagram below shows the architecture blueprint for Research@Scale. The solution offered on the AWS platform covers technology, solutions, and integrated services to help large enterprises manage research across global locations.
Leveraging AWS Biotech Blueprint with our Research Gateway
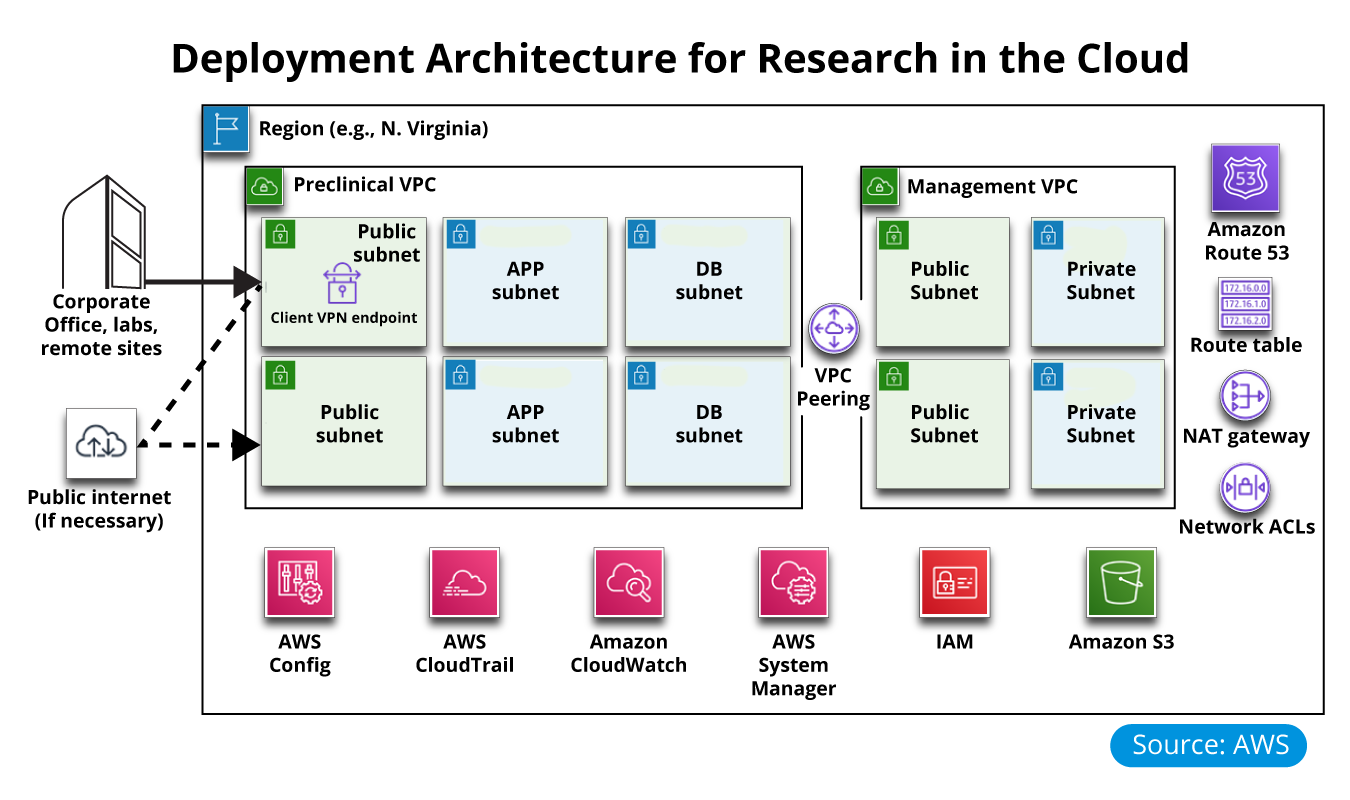
Use case with AWS Biotech Blueprint that provides a Core template for deploying a preclinical, cloud-based research infrastructure and optional informatics software on AWS.
This Quick Start sets up the following:
- A highly available architecture that spans two availability zones
- A preclinical virtual private cloud (VPC) configured with public and private subnets according to AWS best practices to provide you with your own virtual network on AWS. This is where informatics and research applications will run
- A management VPC configured with public and private subnets to support the future addition of IT-centric workloads such as active directory, security appliances, and virtual desktop interfaces
- Redundant, managed NAT gateways to allow outbound internet access for resources in the private subnets
- Certificate-based virtual private network (VPN) services through the use of AWS Client VPN endpoints
- Private, split-horizon Domain Name System (DNS) with Amazon Route 53
- Best-practice AWS Identity and Access Management (IAM) groups and policies based on the separation of duties, designed to follow the U.S. National Institute of Standards and Technology (NIST) guidelines
- A set of automated checks and alerts to notify you when AWS Config detects insecure configurations
- Account-level logging, audit, and storage mechanisms are designed to follow NIST guidelines
- A secure way to remotely join the preclinical VPC network is by using the AWS Client VPN endpoint
- A prepopulated set of AWS Systems Manager Parameter Store key/value pairs for common resource IDs
- (Optional) An AWS Service Catalog portfolio of common informatics software that can be easily deployed into your preclinical VPC
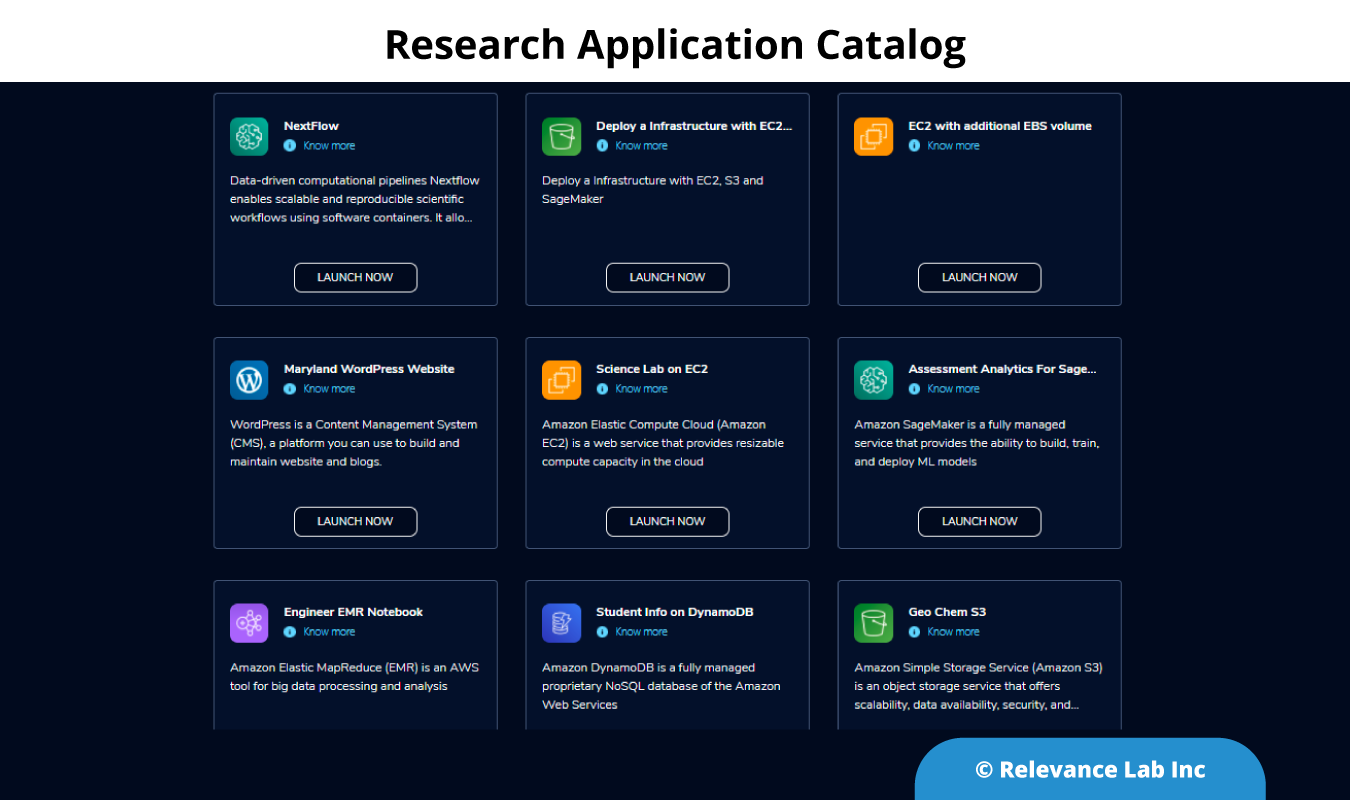
Using the Quickstart templates, the products were added to AWS Service Catalog and imported into RLCatalyst Research Gateway.
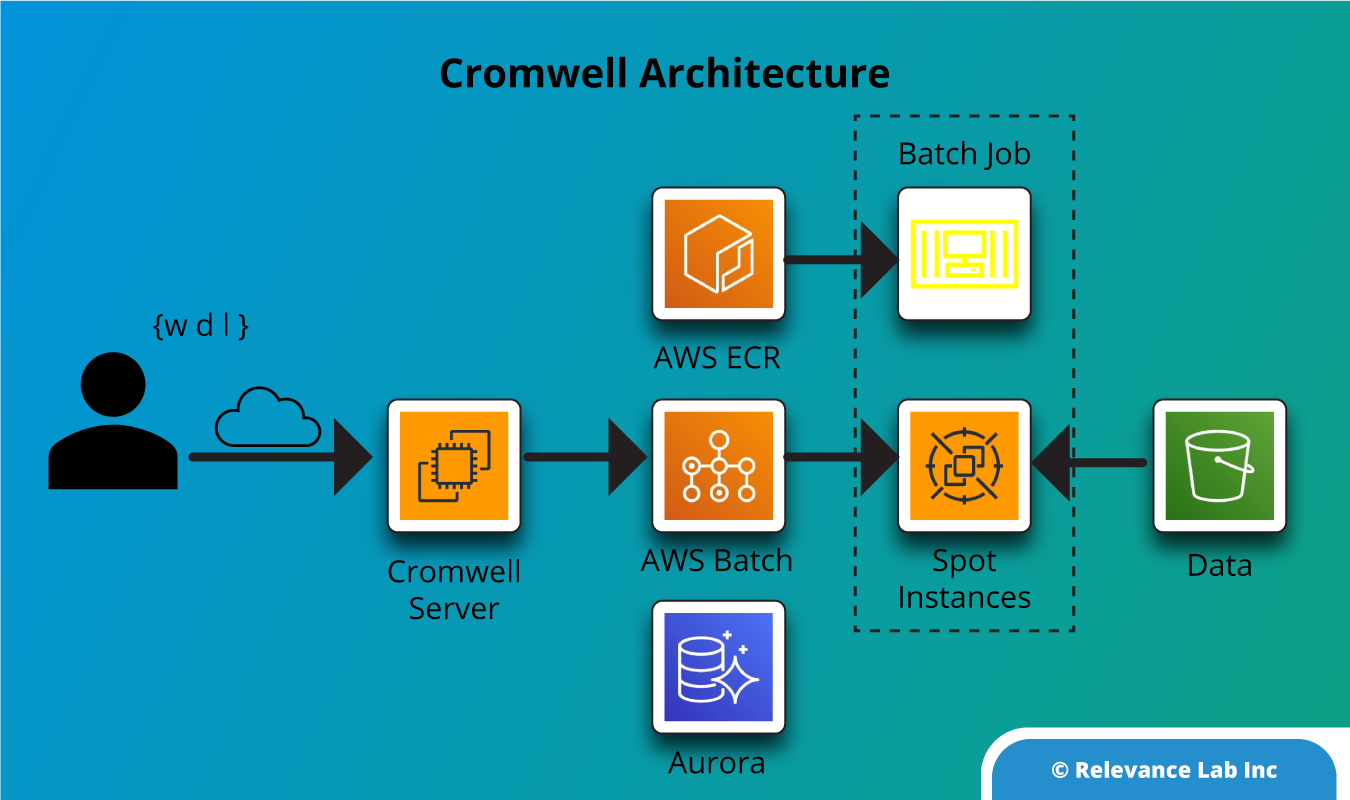
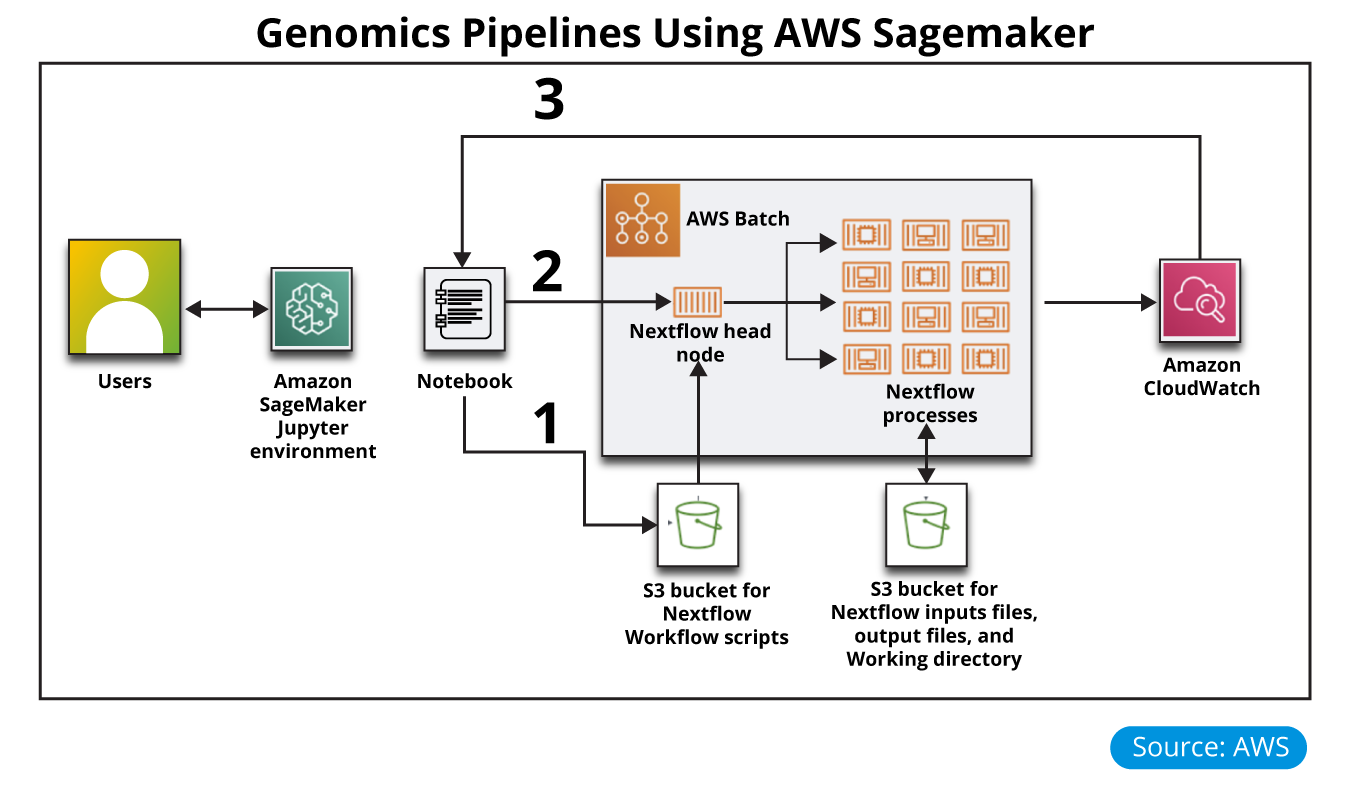
Using the standard products, the Nextflow Workflow Orchestration engine was launched for Genomics pipeline analysis. Nextflow helps to create and orchestrate analysis workflows and AWS Batch to run the workflow processes.
Nextflow is an open-source workflow framework and domain-specific language (DSL) for Linux, developed by the Comparative Bioinformatics group at the Barcelona Centre for Genomic Regulation (CRG). The tool enables you to create complex, data-intensive workflow pipeline scripts, and simplifies the implementation and deployment of genomics analysis workflows in the cloud.
This Quick Start sets up the following environment in a preclinical VPC:
- In the public subnet, an optional Jupyter notebook in Amazon SageMaker is integrated with an AWS Batch environment.
- In the private application subnets, an AWS Batch compute environment for managing Nextflow job definitions and queues and for running Nextflow jobs. AWS Batch containers have Nextflow installed and configured in an Auto Scaling group.
- Because there are no databases required for Nextflow, this Quick Start does not deploy anything into the private database (DB) subnets created by the Biotech Blueprint core Quick Start.
- An Amazon Simple Storage Service (Amazon S3) bucket to store your Nextflow workflow scripts, input and output files, and working directory.
RStudio for Scientific Research
RStudio is a popular IDE, licensed either commercially or under AGPLv3, for working with R. RStudio is available in a desktop version or a server version that allows you to access R via a web browser.
After you’ve analyzed the results, you may want to visualize them. Shiny is a great R package, licensed either commercially or under AGPLv3, that you can use to create interactive dashboards. Shiny provides a web application framework for R. It turns your analyses into interactive web applications; no HTML, CSS, or JavaScript knowledge is required. Shiny Server can deliver your R visualization to your customers via a web browser and execute R functions, including database queries, in the background.
RStudio is provided as a standard catalog item in Research Gateway for 1-Click deployment and use. AWS provides a number of tools like AWS Athena, AWG Glue, and others to connect to datasets for research analysis.
Benefits of using AWS for Clinical Informatics
- Data transfer and storage The volume of genomics data poses challenges for transferring it from sequencers in a quick and controlled fashion, then finding storage resources that can accommodate the scale and performance at a price that is not cost-prohibitive. AWS enables researchers to manage large-scale data that has outpaced the capacity of on-premises infrastructure. By transferring data to the AWS Cloud, organizations can take advantage of high-throughput data ingestion, cost-effective storage options, secure access, and efficient searching to propel genomics research forward.
- Workflow automation for secondary analysis Genomics organizations can struggle with tracking the origins of data when performing secondary analyses and running reproducible and scalable workflows while minimizing IT overhead. AWS offers services for scalable, cost-effective data analysis and simplified orchestration for running and automating parallelizable workflows. Options for automating workflows enable reproducible research or clinical applications, while AWS native, partner (NVIDIA and DRAGEN), and open source solutions (Cromwell and Nextflow) provide flexible options for workflow orchestrators to help scale data analysis.
- Data aggregation and governance Successful genomics research and interpretation often depend on multiple, diverse, multi-modal datasets from large populations. AWS enables organizations to harmonize multi-omic datasets and govern robust data access controls and permissions across a global infrastructure to maintain data integrity as research involves more collaborators and stakeholders. AWS simplifies the ability to store, query, and analyze genomics data, and link with clinical information.
- Interpretation and deep learning for tertiary analysis Analysis requires integrated multi-modal datasets and knowledge bases, intensive computational power, big data analytics, and machine learning at scale, which, historically can take weeks or months, delaying time to insights. AWS accelerates the analysis of big genomics data by leveraging machine learning and high-performance computing. With AWS, researchers have access to greater computing efficiencies at scale, reproducible data processing, data integration capabilities to pull in multi-modal datasets, and public data for clinical annotation—all within a compliance-ready environment.
- Clinical applications There are several hindrances that impede the scale and adoption of genomics for clinical applications including speed of analysis, managing protected health information (PHI), and providing reproducible and interpretable results. By leveraging the capabilities of the AWS Cloud, organizations can establish a differentiated capability in genomics to advance their applications in precision medicine and patient practice. AWS services enable the use of genomics in the clinic by providing the data capture, compute, and storage capabilities needed to empower the modernized clinical lab to decrease the time to results, all while adhering to the most stringent patient privacy regulations.
- Open datasets As more life science researchers move to the cloud and develop cloud-native workflows, they bring reference datasets with them, often in their own personal buckets, leading to duplication, silos, and poor version documentation of commonly used datasets. The AWS Open Data Program (ODP) helps democratize data access by making it readily available in Amazon S3, providing the research community with a single documented source of truth. This increases study reproducibility, stimulates community collaboration, and reduces data duplication. The ODP also covers the cost of Amazon S3 storage, egress, and cross-region transfer for accepted datasets.
- Cost optimization Researchers utilize massive genomics datasets, which require large-scale storage options and powerful computational processing and can be cost-prohibitive. AWS presents cost-saving opportunities for genomics researchers across the data lifecycle—from storage to interpretation. AWS infrastructure and data services enable organizations to save time, money, and devote more resources to science.
Summary
Relevance Lab is a specialist AWS partner working closely in Health Informatics and Genomics solutions leveraging AWS existing solutions and complementing them with its Self-Service Cloud Portal solutions, automation, and governance best practices.
To know more about how we can help standardize, scale, and speed up Scientific Research in Cloud, feel free to contact us at marketing@relevancelab.com.
References
AWS Whitepaper on Genomics Data Transfer, Analytics and Machine Learning
Genomics Workflows on AWS
HPC on AWS Video – Running Genomics Workflows with Nextflow
Workflow Orchestration with Nextflow on AWS Cloud
Biotech Blueprint on AWS Cloud
Running R on AWS
Advanced Bioinformatics Workshop