2022 Blogs, Blog, Featured
Complex Genomics Analysis Pipelines made Simple with NextFlow & Research Gateway integrated with Cost Tracking and Security
As a researcher, do you want to get started in minutes to run any complex genomics pipeline with large data sets without worrying about hours to set up the environment, dealing with large data sets availability & storage, security of your cloud infrastructure, and most of all unknown expenses? RLCatalyst makes your life simpler, and in this blog, we will cover how easy it is to use publicly available Genomics pipelines from nf-co.re using Nextflow on your AWS Cloud environment with ease.
There are a number of open-source tools available for researchers driving re-use. However, what Research Institutions and Genomics companies are looking for is a right balance on three key dimensions before adopting cloud in a large scale manner for internal use:
- Cost and Budget Governance: Strong focus on Cost Tracking of Cloud resources to track, analyze, control, and optimize budget spends.
- Research Data & Tools Easy Collaboration: Principal Investigators and researchers need to focus on data management, governance, and privacy along with analysis and collaboration in real-time without worrying about Cloud complexity.
- Security and Compliance: Research requires a strong focus on security and compliance covering Identity management, data privacy, audit trails, encryption, and access management.
To make sure the above functionalities do not slow down researchers from focussing on Science due to complexities of infrastructure, Research Gateway provides the reliable solution by automating cost & budget tracking with safe-guards and providing a simple self-service model for collaboration. We will demonstrate in this blog how researchers can use a vast set of publicly available tools, pipelines and data easily on this platform with tight budget controls.
Here is a quick video of the ease with which researchers can get started in a frictionless manner.
nf-co.re is a community effort to collect a curated set of analysis pipelines built using Nextflow. The key aspects of these pipelines are that these pipelines adhere to strict guidelines that ensure they can be reused extensively. These pipelines have following advantages:
- Cloud-Ready – Pipelines are tested on AWS after every release. You can even browse results live on the website and use outputs for your own benchmarking.
- Portable and reproducible – Pipelines follow best practices to ensure maximum portability and reproducibility. The large community makes the pipelines exceptionally well tested and easy to run.
- Packaged software – Pipeline dependencies are automatically downloaded and handled using Docker, Singularity, Conda, or others. No need for any software installations.
- Stable releases – nf-core pipelines use GitHub releases to tag stable versions of the code and software, making pipeline runs totally reproducible.
- CI testing – Every time a change is made to the pipeline code, nf-core pipelines use continuous integration testing to ensure that nothing has broken.
- Documentation – Extensive documentation covering installation, usage, and description of output files ensures that you won’t be left in the dark.
Sample of commonly used pipelines that are supported out-of-box in Research Gateway to run with a few clicks and do important genomic analysis. While publicly available repos are easily accessible, it also allows private repositories and custom pipelines to run with ease.
| Pipeline Name | Description | Commonly used for |
|---|---|---|
| Sarek | Analysis pipeline to detect germline or somatic variants (pre-processing, variant calling, and annotation) from Whole Genome Sequencing (WGS) / targeted sequencing | Variant Analysis – workflow designed to detect variants on whole genome or targeted sequencing data |
| RNA-Seq | RNA-Sequencing analysis pipeline using STAR, RSEM, HISAT2, or Salmon with gene/isoform counts and extensive quality control | Common basic analysis for RNA-Sequencing with a reference genome and annotation |
| Dual RNA-Seq | Analysis of Dual RNA-Seq data – an experimental method for interrogating host-pathogen interactions through simultaneous RNA-Seq | Specifically used for the analysis of Dual RNA-Seq data, interrogating host-pathogen interactions through simultaneous RNA-Seq |
| Bactopia | Bactopia is a flexible pipeline for complete analysis of bacterial genomes | Bacterial Genomic Analysis with focus on Food Safety |
| Viralrecon | Assembly and intrahost/low-frequency variant calling for viral samples | Supports metagenomics and amplicon sequencing data derived from the Illumina sequencing platform |
*The above samples can be launched in less than 5 min and take less than $5 to run with test data and 80% productivity gains achieved.
The figure below shows the building block of this solution on AWS Cloud.
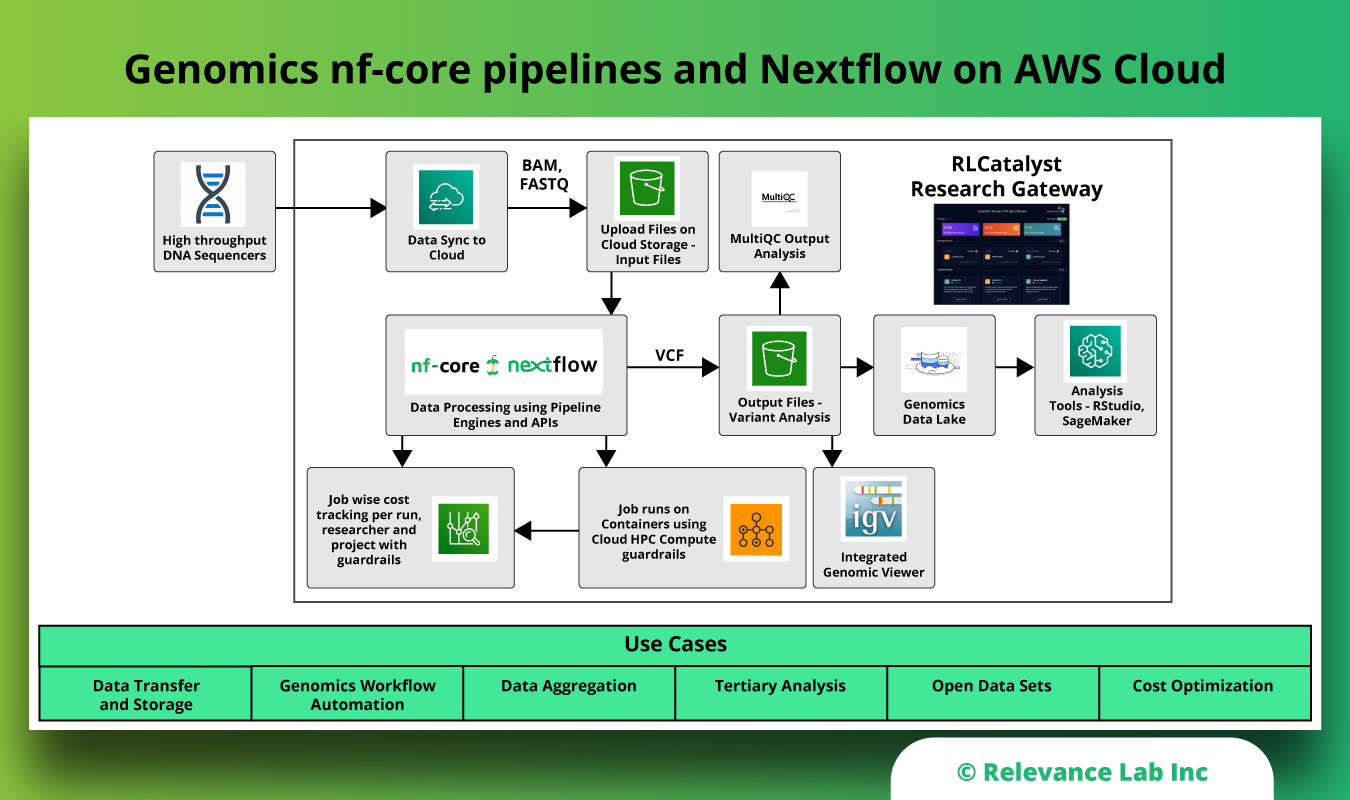
Steps for running nf-core pipeline with Nextflow on AWS Cloud
| Steps | Details | Time Taken |
|---|---|---|
| 1. | Log into RLCatalyst Research Gateway as a Principal Investigator or Researcher profile. Select the project for running Genomics Pipelines, and first time create a new Nextflow Advanced Product. | 5 min |
| 2. | Select the Input Data location, output data location, pipeline to run (from nf-co.re), and provide parameters (container path, data pattern to use, etc.). Default parameters are already suggested for use of AWS Batch with Spot instances and all other AWS complexities abstracted from end-user for simplicity. | 5 min to provision new Nextflow & Nextflow Tower Server on AWS with AWS Batch setup completed with 1-Click |
| 3. | Execute Pipeline (using UI interface or by SSH into Head-node) on Nextflow Server. There is ability to run the new pipelines, monitor status, and review outputs from within the Portal UI. | Pipelines can take some time to run depending on the size of data and complexity |
| 4. | Monitor live pipelines with the 1-Click launch of Nextflow Tower integrated with the portal. Also, view outputs of the pipeline in outputs S3 bucket from within the Portal. Use specialized tools like MultiQC, IGV, and RStudio for further analysis. | 5 min |
| 5. | All costs related to User, Product, and Pipelines are automatically tagged and can be viewed in the Budgets screen to know the Cloud spend for pipeline execution that includes all resources, including AWS Batch HPC instances dynamically provisioned. Once the pipelines are executed, the existing Cromwell Server can be stopped or terminated to reduce ongoing costs. | 5 min |
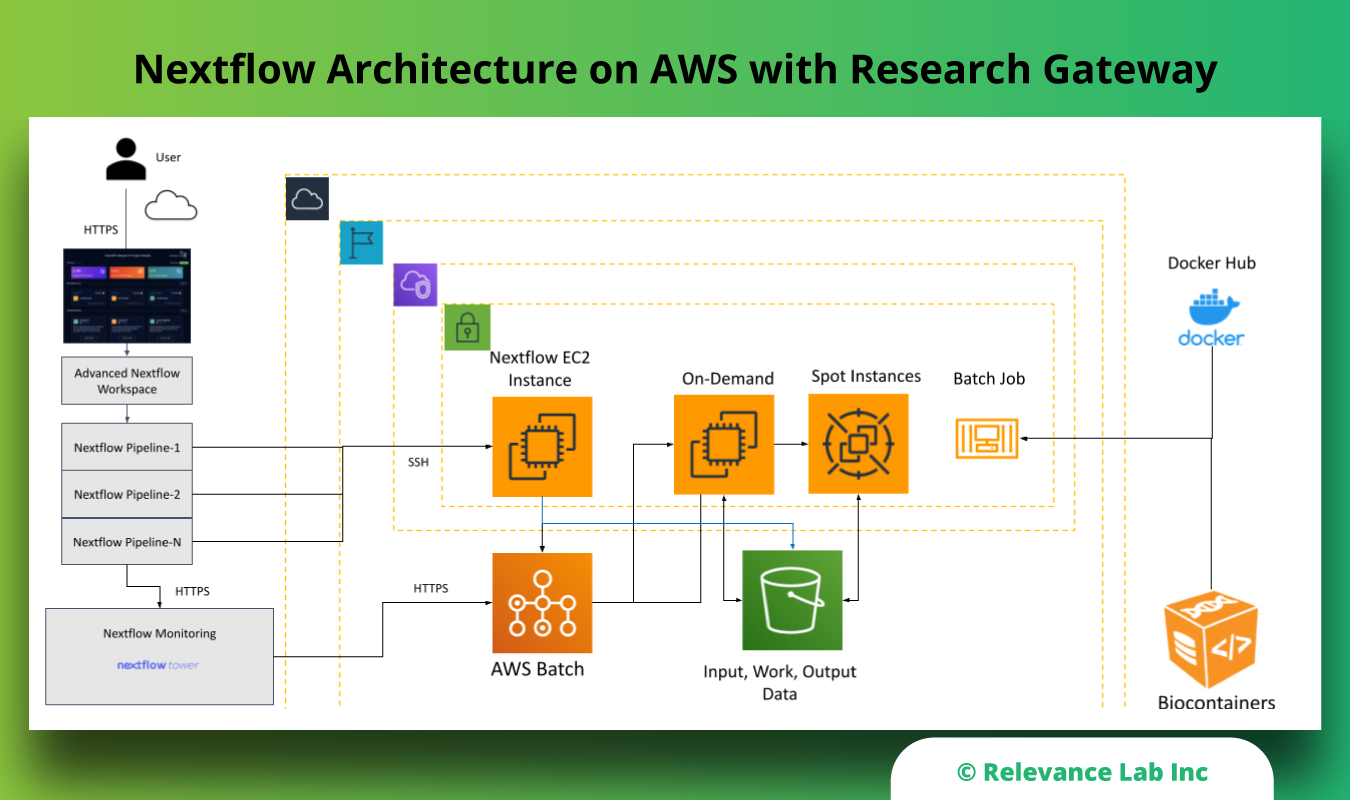
The figure below shows the Nextflow Architecture on AWS.
Summary
nf-co.re community is constantly striving to make Genomics Research in the Cloud simpler. While these pipelines are easily available, running them on AWS Cloud with proper cost tracking, collaboration, data management, and integrated workbench were missing that is now solved by Research Gateway. Relevance Lab, in partnership with AWS, has addressed this need with their Genomics Cloud solution to make scientific research frictionless.
To know more about how you can start your Nextflow nf-co.re pipelines on the AWS Cloud in 30 minutes using our solution at https://research.rlcatalyst.com, feel free to contact marketing@relevancelab.com
References
Enabling Researchers with Next-Generation Sequencing (NGS) Leveraging Nextflow and AWS
Pipelining GATK with WDL and Cromwell on AWS Cloud
Genomics Cloud on AWS with RLCatalyst Research Gateway
Health Informatics and Genomics on AWS with RLCatalyst Research Gateway
Accelerating Genomics and High Performance Computing on AWS with Relevance Lab Research Gateway Solution
